-Search query
-Search result
Showing 1 - 50 of 341 items for (author: miller & as)

EMDB-41126: 
Cryo-EM structure of the human CLC-2 chloride channel transmembrane domain with bound inhibitor AK-42
Method: single particle / : Xu M, Neelands T, Powers AS, Liu Y, Miller S, Pintilie G, Du Bois J, Dror RO, Chiu W, Maduke M

EMDB-41127: 
Cryo-EM structure of the human CLC-2 chloride channel transmembrane domain Apo state with resolved N-terminal hairpin
Method: single particle / : Xu M, Neelands T, Powers AS, Liu Y, Miller S, Pintilie G, Du Bois J, Dror RO, Chiu W, Maduke M

EMDB-41128: 
Cryo-EM structure of the human CLC-2 chloride channel transmembrane domain with symmetric C-terminal
Method: single particle / : Xu M, Neelands T, Powers AS, Liu Y, Miller S, Pintilie G, Du Bois J, Dror RO, Chiu W, Maduke M

EMDB-41129: 
Title: Cryo-EM structure of the human CLC-2 chloride channel transmembrane domain with asymmetric C-terminal
Method: single particle / : Xu M, Neelands T, Powers AS, Liu Y, Miller S, Pintilie G, Du Bois J, Dror RO, Chiu W, Maduke M

EMDB-41130: 
Cryo-EM structure of the human CLC-2 chloride channel C-terminal domain
Method: single particle / : Xu M, Neelands T, Powers AS, Liu Y, Miller S, Pintilie G, Du Bois J, Dror RO, Chiu W, Maduke M

PDB-8t1n: 
Micro-ED Structure of a Novel Domain of Unknown Function Solved with AlphaFold
Method: electron crystallography / : Miller JE, Cascio D, Sawaya MR, Cannon KA, Rodriguez JA, Yeates TO

EMDB-40856: 
Single particle reconstruction of the human LINE-1 ORF2p without substrate (apo)
Method: single particle / : van Eeuwen T, Taylor MS, Rout MP
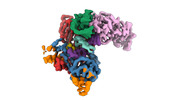
EMDB-40858: 
Structure of LINE-1 ORF2p with template:primer hybrid
Method: single particle / : van Eeuwen T, Taylor MS, Rout MP
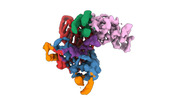
EMDB-40859: 
Structure of LINE-1 ORF2p with an oligo(A) template
Method: single particle / : van Eeuwen T, Taylor MS, Rout MP

PDB-8sxt: 
Structure of LINE-1 ORF2p with template:primer hybrid
Method: single particle / : van Eeuwen T, Taylor MS, Rout MP

PDB-8sxu: 
Structure of LINE-1 ORF2p with an oligo(A) template
Method: single particle / : van Eeuwen T, Taylor MS, Rout MP

EMDB-16005: 
GABA-A receptor a5 homomer - a5V3 - APO
Method: single particle / : Miller PS, Malinauskas TM, Hardwick SW, Chirgadze DY

EMDB-16050: 
GABA-A receptor a5 homomer - a5V3 - Basmisanil - HR
Method: single particle / : Malinauskas TM, Wahid AA, Hardwick SW, Chirgadze DY, Miller PS

EMDB-16051: 
GABA-A receptor a5 homomer - a5V3 - RO154513
Method: single particle / : Miller PS, Malinauskas TM, Hardwick SW, Chirgadze DY, Wahid AA

EMDB-16055: 
GABA-A receptor a5 homomer - a5V3 - RO5211223
Method: single particle / : Miller PS, Malinauskas TM, Hardwick SW, Chirgadze DY

EMDB-16058: 
GABA-A receptor a5 homomer - a5V3 - Diazepam
Method: single particle / : Miller PS, Malinauskas TM, Kasaragod VB, Chirgadze DY

EMDB-16060: 
GABA-A receptor a5 homomer - a5V3 - DMCM
Method: single particle / : Miller PS, Malinauskas TM, Hardwick SW, Chirgadze DY

EMDB-16063: 
GABA-A receptor a5 homomer - a5V3 - L655708
Method: single particle / : Miller PS, Malinauskas TM, Hardwick SW, Chirgadze DY

EMDB-16066: 
GABA-A receptor a5 homomer - a5V3 - RO7172670
Method: single particle / : Miller PS, Malinauskas TM, Hardwick SW, Kasaragod VB

EMDB-16067: 
GABA-A receptor a5 homomer - a5V3 - RO7015738
Method: single particle / : Miller PS, Malinauskas TM, Kasaragod VB, Chirgadze DY

EMDB-16068: 
GABA-A receptor a5 homomer - a5V3 - RO4938581
Method: single particle / : Miller PS, Malinauskas TM, Hardwick SW, Chirgadze DY

PDB-8bej: 
GABA-A receptor a5 homomer - a5V3 - APO
Method: single particle / : Miller PS, Malinauskas TM, Hardwick SW, Chirgadze DY

PDB-8bha: 
GABA-A receptor a5 homomer - a5V3 - Basmisanil - HR
Method: single particle / : Malinauskas TM, Wahid AA, Hardwick SW, Chirgadze DY, Miller PS

PDB-8bhb: 
GABA-A receptor a5 homomer - a5V3 - RO154513
Method: single particle / : Miller PS, Malinauskas TM, Hardwick SW, Chirgadze DY, Wahid AA

PDB-8bhi: 
GABA-A receptor a5 homomer - a5V3 - RO5211223
Method: single particle / : Miller PS, Malinauskas TM, Hardwick SW, Chirgadze DY

PDB-8bhk: 
GABA-A receptor a5 homomer - a5V3 - Diazepam
Method: single particle / : Miller PS, Malinauskas TM, Kasaragod VB, Chirgadze DY

PDB-8bhm: 
GABA-A receptor a5 homomer - a5V3 - DMCM
Method: single particle / : Miller PS, Malinauskas TM, Hardwick SW, Chirgadze DY

PDB-8bho: 
GABA-A receptor a5 homomer - a5V3 - L655708
Method: single particle / : Miller PS, Malinauskas TM, Hardwick SW, Chirgadze DY

PDB-8bhq: 
GABA-A receptor a5 homomer - a5V3 - RO7172670
Method: single particle / : Miller PS, Malinauskas TM, Hardwick SW, Kasaragod VB

PDB-8bhr: 
GABA-A receptor a5 homomer - a5V3 - RO7015738
Method: single particle / : Miller PS, Malinauskas TM, Kasaragod VB

PDB-8bhs: 
GABA-A receptor a5 homomer - a5V3 - RO4938581
Method: single particle / : Miller PS, Malinauskas TM, Hardwick SW, Chirgadze DY

EMDB-29452: 
Structure Of Respiratory Syncytial Virus Polymerase with Novel Non-Nucleoside Inhibitor
Method: single particle / : Yu X, Abeywickrema P, Bonneux B, Behera I, Jacoby E, Fung A, Adhikary S, Bhaumik A, Carbajo RJ, Bruyn SD, Miller R, Patrick A, Pham Q, Piassek M, Verheyen N, Shareef A, Sutto-Ortiz P, Ysebaert N, Vlijmen HV, Jonckers THM, Herschke F, McLellan JS, Decroly E, Fearns R, Grosse S, Roymans D, Sharma S, Rigaux P, Jin Z

PDB-8fu3: 
Structure Of Respiratory Syncytial Virus Polymerase with Novel Non-Nucleoside Inhibitor
Method: single particle / : Yu X, Abeywickrema P, Bonneux B, Behera I, Jacoby E, Fung A, Adhikary S, Bhaumik A, Carbajo RJ, Bruyn SD, Miller R, Patrick A, Pham Q, Piassek M, Verheyen N, Shareef A, Sutto-Ortiz P, Ysebaert N, Vlijmen HV, Jonckers THM, Herschke F, McLellan JS, Decroly E, Fearns R, Grosse S, Roymans D, Sharma S, Rigaux P, Jin Z

EMDB-29281: 
Cryo-EM structure of STING oligomer bound to cGAMP and NVS-STG2
Method: single particle / : Li J, Canham SM, Zhang X, Bai X, Feng Y

EMDB-29282: 
Cryo-EM structure of STING oligomer bound to cGAMP, NVS-STG2 and C53
Method: single particle / : Li J, Canham SM, Zhang X, Bai X, Feng Y

PDB-8flk: 
Cryo-EM structure of STING oligomer bound to cGAMP and NVS-STG2
Method: single particle / : Li J, Canham SM, Zhang X, Bai X, Feng Y

PDB-8flm: 
Cryo-EM structure of STING oligomer bound to cGAMP, NVS-STG2 and C53
Method: single particle / : Li J, Canham SM, Zhang X, Bai X, Feng Y

EMDB-17540: 
Cryo-EM structure of full-length human UBR5 (homotetramer)
Method: single particle / : Aguirre JD, Kater L, Kempf G, Cavadini S, Thoma NH

PDB-8p83: 
Cryo-EM structure of full-length human UBR5 (homotetramer)
Method: single particle / : Aguirre JD, Kater L, Kempf G, Cavadini S, Thoma NH

EMDB-28731: 
Dimeric complex of DNA-PKcs
Method: single particle / : Chen S, He Y

EMDB-28732: 
NHEJ Long-range complex with PAXX
Method: single particle / : Chen S, He Y

EMDB-28733: 
NHEJ Long-range complex with ATP
Method: single particle / : Chen S, He Y

EMDB-28734: 
DNA-PKcs monomer in dimer DNA-PKcs complex
Method: single particle / : Chen S, He Y

EMDB-28735: 
DNAPKcs-Ku-LigIV(BRCT)-PAXX-DNA subcomplex in PAXX-LR complex
Method: single particle / : Chen S, He Y
EMDB-28736: 
XRCC4-XLF-XRCC4 subcomplex of PAXX-LR complex
Method: single particle / : Chen S, He Y

EMDB-28737: 
DNA-PKcs without N-terminus in PAXX-LR-ATP complex
Method: single particle / : Chen S, He Y

EMDB-28738: 
DNAPKcs-Ku-DNA subcomplex in PAXX-LR-ATP complex
Method: single particle / : Chen S, He Y

EMDB-28739: 
XRCC4-XLF-XRCC4 subcomplex in LR-ATP complex
Method: single particle / : Chen S, He Y
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search





 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
